Aggregation Prone Regions (APRs)
The short-stretch hypothesis of protein aggregation
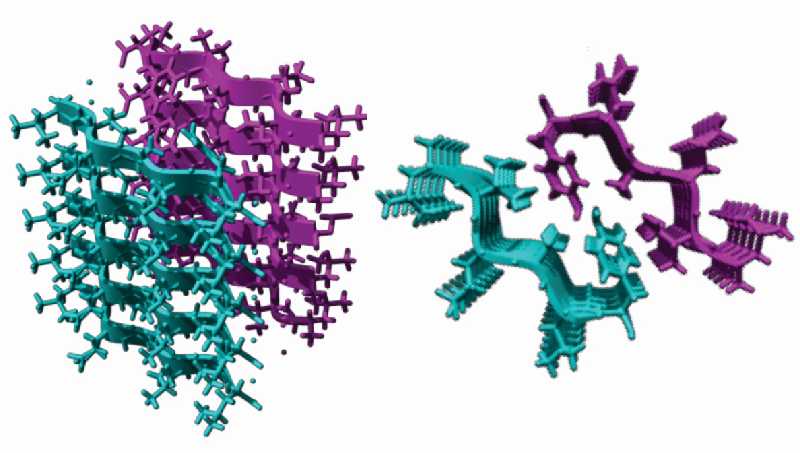
Aggregation Prone Regions (APRs) are 5-15 residue long stretches in proteins that have a tendency to self-associate into ordered intermolecular beta-sheet arrangements called “cross-beta” spines [1]. In this conformation, APRs are stapled together into beta-sheets through backbone hydrogen bonds and these beta-sheets pack in layers. The layers form stable structures, known as “steric zippers”, because the amino acid side chains of the layers tightly inter-digitate with the side chains from the neighbouring layers (see Figure).
The “cross-beta” spine structure of amyloid fibrils. The tightly fitting side chains from the teal and the purple coloured beta-sheets form a “steric zipper”. The two sides of the figure are two views of the same structure, rotated by 90°.
APRs are mainly hydrophobic; however, other structural properties such as beta-propensity, sidechain size and charge are equally important for accurate prediction of aggregation propensity [2,3]. Moreover, the real aggregation potential is also related to solvent accessibility of APRs, supporting the notion that the rest of the protein can also contribute to self-assembly without being a core part of the ultimate fibril structure.
Most proteins possess at least one APR, but APRs usually do not induce aggregation because they are buried inside the hydrophobic core of the folded protein. The self-assembly of APRs can drive protein aggregation only in situations where proteins are partially or completely unfolded, and the APRs are exposed, e.g. during protein translation or translocation, during transient protein breathing motions, under situations of physiological stress or due to mutations that destabilise the native conformation. APRs are also major determinants during folding (e.g. co-translational aggregation), where elevated expression and low elongation rates can shift the scale between folding and aggregation towards the latter.
APRs are necessary and sufficient for driving protein aggregation: grafting APRs onto non-aggregating proteins drives them to form ordered amyloid-like fibrils. The decisive role of APRs in aggregation is further supported by co-expression studies of unrelated aggregating peptides sharing no sequence similarity, which results in the formation of separate inclusion bodies in mammalian, as well as E. coli cells. Finally, sequence dependence of aggregation interactions is also pinpointed by in vitro aggregation reactions, where only closely related aggregation prone sequences are capable of effectively seeding nucleation-dependent kinetics.
Combined with the structural restraints of the “cross-beta" conformation, the above suggest that protein aggregation is not simply the result of a generalized hydrophobic collapse of misfolded proteins but rather the required outcome of highly specific, possibly sequence- and structure dependent interactions between aggregation prone amino acid stretches.
References
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 22 (10):1302-1306. doi:10.1038/nbt1012
- De Baets G, Schymkowitz J, Rousseau F (2014) Predicting aggregation-prone sequences in proteins. In: Perrett S (ed) Amyloids in Health and Disease, vol 56. Essays in Biochemistry. Portland Press Ltd, London, pp 41-52. doi:10.1042/bse0560041
- Louros N, Orlando G, De Vleeshouwer, M; Rousseau, F; Schymkowitz, J (2020) Structure-based machine-guided mapping of amyloid sequence space reveals uncharted sequence clusters with higher solubilities. Nature Communication