Gatekeepers (GKs)
Evolutionary selection of gatekeeper residues to control aggregation
Most proteins across the evolutionary spectrum from bacteria through plants to mammals contain at least one and usually several aggregation prone regions (APRs) [1]. Since protein aggregation is detrimental to the function of most proteins and APRs promote protein aggregation, the question arises why Evolution hasn’t eliminated APRs?
It turns out that some of the forces that drive protein aggregation, i.e. beta-sheet formation and hydrophobicity, are also crucial for proper protein folding [2]. Hence, most stable globular folds contain APRs. Moreover, the genetic code ensures that once APRs are part of a polypeptide chain, they are “locked-in” [3]. Since APRs are unavoidable and conserved, Evolution implemented several limitations on them to reduce the threat they pose. The so-called aggregation gatekeepers (GKs) are one of these measures.
The amino acid residues arginine (Arg, R), lysine (Lys, K), aspartic acid (Asp D) and glutamic acid (Glu, E), as well as proline (Pro, P), are significantly enriched in the regions directly flanking APRs [1,4]. These gatekeeper residues (GKs) impede aggregation in several ways: First, charged residues such as Arg, Lys, Asp and Glu introduce repulsive forces that prevent aggregation. Second, Arg and Lys further prevent aggregation through entropic costs: it is unfavourable to fix these flexible side chains into one defined position. Furthermore, residues that are incompatible with the beta-structure such as proline (and to a lesser extent, glycine) also contribute to controlling aggregation.
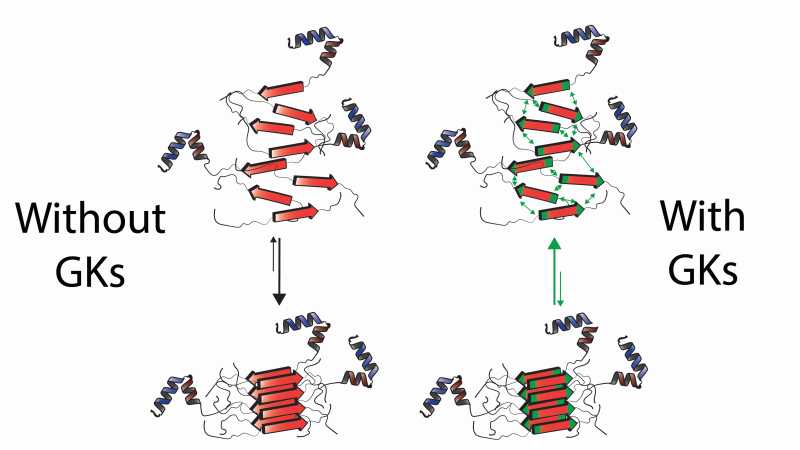
Protein aggregation in the absence and presence of aggregation Gatekeepers (GKs). Unfolded or misfolded proteins containing APRs (red stretches) tend to form intermolecular cross-beta structures known as protein aggregates. These structures are highly stable and generally preclude normal protein function. Gatekeeper residues (green) at the flanks of APRs reduce aggregation propensity through charge repulsion (indicated by green arrows) and their general incompatibility with the cross-beta structure. In doing so, GKs prevent proteins from engaging in these highly ordered structures, shifting the balance away from aggregation, and increasing the chances of proteins acquiring and maintaining their native fold.
Since their discovery, research from the Switch lab has increased our understanding of GK function and Evolution. We showed, for example, that GKs are subject to evolutionary pressure: the enrichment of GKs is more pronounced in long-living than in short-living proteins [2]. Furthermore, a correlation exists between the strength of an APR and the number of GKs flanking it [5]. Introducing GKs helps to make proteins easier to produce and potentially safer to employ as therapeutics [6].The opposite is also true: mutations that remove GK residues are associated with human disease [7], showing that GKs are essential for maintaining protein homeostasis (proteostasis). Indeed, GKs affect not only aggregation but also protein synthesis and degradation rates, heat stress response and overall cellular fitness [8,4]. Furthermore, our lab showed that GKs are conserved despite their structurally destabilizing character (they reduce thermodynamic stability of the native structure by 0,5 kcal/mol on average). The latter observation shows that GKs are indeed a distinct functional class of residues dedicated to aggregation prevention.
In our most recent work on this topic, we focused on the close relationship between GKs and the proteostatic machinery. We showed that whereas acidic GKs are fully autonomous in preventing aggregation, basic GKs rely heavily on the assistance of molecular chaperones. Structural constraints, namely incompatibility with burial into the hydrophobic core of globular proteins, prevent acidic GKs from being used ubiquitously. Our results, therefore, suggest that the use of the inferior basic GKs was necessary for the bigger and more complex globular protein folds. As a compensatory mechanism, molecular chaperones developed a preference for APRs flanked by basic residues, while leaving APRs protected by acidic GKs to fend for themselves [9]. Our work has thus uncovered a triaging mechanism that allows the proteostatic machinery to specifically home in on the most dangerous regions in protein sequences through the GKs that flank them.
References
- Rousseau F, Serrano L, Schymkowitz JW (2006) How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol 355 (5):1037-1047. doi:10.1016/j.jmb.2005.11.035
- De Baets G, Reumers J, Delgado Blanco J, Dopazo J, Schymkowitz J, Rousseau F (2011) An evolutionary trade-off between protein turnover rate and protein aggregation favors a higher aggregation propensity in fast degrading proteins. PLoS Comput Biol 7 (6):e1002090. doi:10.1371/journal.pcbi.1002090
- Langenberg T, Gallardo R, van der Kant R, Louros N, Michiels E, Duran-Romaña R, Houben B, Cassio R, Wilkinson H, Garcia T, Ulens C, Van Durme J, Rousseau F, Schymkowitz J (2020) Thermodynamic and Evolutionary Coupling between the Native and Amyloid State of Globular Proteins. Cell reports 31 (2). doi:10.1016/j.celrep.2020.03.076
- Beerten J, Jonckheere W, Rudyak S, Xu J, Wilkinson H, De Smet F, Schymkowitz J, Rousseau F (2012) Aggregation gatekeepers modulate protein homeostasis of aggregating sequences and affect bacterial fitness. Protein engineering, design & selection : PEDS 25 (7):357-366. doi:10.1093/protein/gzs031
- Reumers J, Maurer-Stroh S, Schymkowitz J, Rousseau F (2009) Protein sequences encode safeguards against aggregation. Hum Mutat 30 (3):431-437. doi:10.1002/humu.20905
- Ganesan A, Siekierska A, Beerten J, Brams M, Van Durme J, De Baets G, Van der Kant R, Gallardo R, Ramakers M, Langenberg T, Wilkinson H, De Smet F, Ulens C, Rousseau F, Schymkowitz J (2016) Structural hot spots for the solubility of globular proteins. Nat Commun 7:10816. doi:10.1038/ncomms10816
- De Baets G, Van Doorn L, Rousseau F, Schymkowitz J (2015) Increased Aggregation Is More Frequently Associated to Human Disease-Associated Mutations Than to Neutral Polymorphisms. PLoS Comput Biol 11 (9):e1004374. doi:10.1371/journal.pcbi.1004374
- Beerten J, Schymkowitz J, Rousseau F (2012) Aggregation prone regions and gatekeeping residues in protein sequences. Curr Top Med Chem 12 (22):2470-2478. doi:10.2174/1568026611212220003
- Houben B, Michiels E, Ramakers M, Konstantoulea K, Louros N, Verniers J, van der Kant R, De Vleeschouwer M, Chicoria N, Vanpoucke T, Gallardo R, Schymkowitz J, Rousseau F (2020) Autonomous aggregation suppression by acidic residues explains why chaperones favour basic residues. EMBO J 39 (11):e102864. doi:10.15252/embj.2019102864