Targeted Protein Aggregation
Using APRs as ZIP codes to target proteins
The amyloid-like aggregation of proteins is driven by short amino acid stretches, called aggregation prone regions (APRs), within the protein sequence. APRs drive intermolecular interactions of proteins by self-assembling into stable structures called “steric zippers”. We have shown in vivo that supplying short amyloidogenic protein fragments (synthetic APRs) can cause functional knockdown of protein(s) by triggering their aggregation, throughout different kingdoms of life (viruses, bacteria, plants, and mammalian cells). Most proteins possess at least one APR, and most APRs are unique in their genome; therefore, APRs can be used to target one specific protein. The Switch laboratory has extensively documented that endogenously expressed proteins that contain amyloidogenic segments (APRs) can be induced to aggregate by seeding their aggregation with a peptide comprising the protein’s own amyloidogenic fragment, even if the proteins are not known to do so either under normal or pathological conditions. We have developed short amyloidogenic protein fragments for the selective suppression of several protein targets in Arabidopsis and maize, resulting in transgenic plants displaying phenotypes such as increased growth (see figure below) or increased starch content [1,2].
Arabidopsis plants expressing an amyloidogenic peptide that knocks down a negative regulator of brassinosteroids (right) grow larger than control plants (left). Figure adapted from [1].
Subsequently, we designed an anti-tumoral amyloidogenic peptide targeting an APR located in the human vascular endothelial growth factor receptor 2 (VEGFR2). This peptide induced the aggregation of VEGFR2, thereby knocking down its function and reducing VEGFR2-dependent growth of tumour allografts of the mouse B16 melanoma line [3]. The amyloidogenic peptide specifically killed tumour cells only that relied on VEGFR2 signalling for survival, without harming other human cells. We have also created synthetic amyloidogenic peptides against the proteome of Staphylococcus aureus (Gram-positive bacteria). These anti-bacterial peptides cured bacterial sepsis caused by methicillin-resistant Staphylococcus aureus in mice without any apparent toxic side-effects [4]. We demonstrated that by targeting the minority of APRs that are redundant in a genome, it is possible to cause the co-aggregation of many proteins, causing the collapse of protein homeostasis (proteostasis). This proved to be an effective way to kill Gram-negative bacteria that are notoriously hard to target. Amyloidogenic peptides targeting redundant APRs induced the formation of inclusion bodies and subsequently, the death of drug-resistant clinical strains of Escherichia coli and Acinetobacter baumannii bacteria and reduced bacterial load in a murine bladder infection model [5]. Most recently, we showed that the targeted protein aggregation approach is highly effective against viruses, as well. As viral replication requires the production of high amounts of viral proteins, proper protein folding is an Achilles heel for the virus. Short amyloidogenic protein fragments that target influenza A and Zika virus stopped the replication of these viruses by causing aggregation of the viral proteins in infected cells [6] (see figure below).
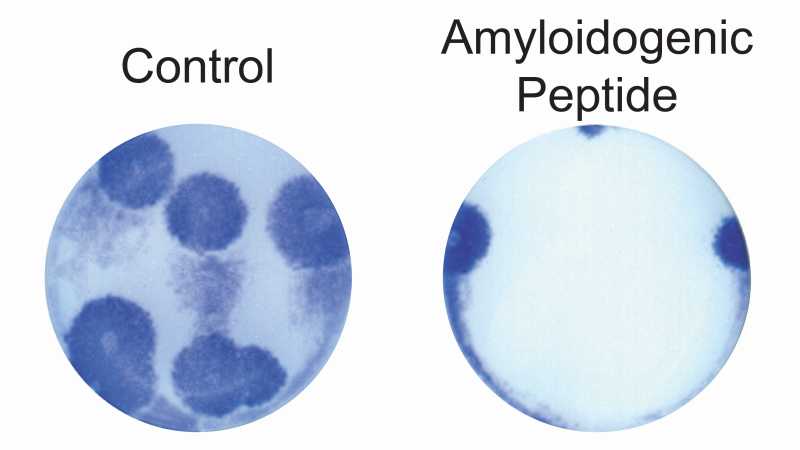
MDCK cells treated with an amyloidogenic anti-influenza peptide (right side) prior to infection with influenza A/PR8 developed fewer and smaller viral plaques than control cells (left side). Figure adapted from [6].
References
- Betti C, Vanhoutte I, Coutuer S, De Rycke R, Mishev K, Vuylsteke M, Aesaert S, Rombaut D, Gallardo R, De Smet F, Xu J, Van Lijsebettens M, Van Breusegem F, Inze D, Rousseau F, Schymkowitz J, Russinova E (2016) Sequence-Specific Protein Aggregation Generates Defined Protein Knockdowns in Plants. Plant physiology 171 (2):773-787. doi:10.1104/pp.16.00335
- Betti C, Schymkowitz J, Rousseau F, Russinova E (2018) Selective Knockdowns in Maize by Sequence-Specific Protein Aggregation. In: Lagrimini LM (ed) Maize: Methods and Protocols, vol 1676. Methods in Molecular Biology. Humana Press Inc, Totowa, pp 109-127. doi:10.1007/978-1-4939-7315-6_6
- Gallardo R, Ramakers M, De Smet F, Claes F, Khodaparast L, Khodaparast L, Couceiro JR, Langenberg T, Siemons M, Nystrom S, Young LJ, Laine RF, Young L, Radaelli E, Benilova I, Kumar M, Staes A, Desager M, Beerens M, Vandervoort P, Luttun A, Gevaert K, Bormans G, Dewerchin M, Van Eldere J, Carmeliet P, Vande Velde G, Verfaillie C, Kaminski CF, De Strooper B, Hammarstrom P, Nilsson KP, Serpell L, Schymkowitz J, Rousseau F (2016) De novo design of a biologically active amyloid. Science 354 (6313). doi:10.1126/science.aah4949
- Bednarska NG, van Eldere J, Gallardo R, Ganesan A, Ramakers M, Vogel I, Baatsen P, Staes A, Goethals M, Hammarstrom P, Nilsson KP, Gevaert K, Schymkowitz J, Rousseau F (2016) Protein aggregation as an antibiotic design strategy. Mol Microbiol 99 (5):849-865. doi:10.1111/mmi.13269
- Khodaparast L, Khodaparast L, Gallardo R, Louros NN, Michiels E, Ramakrishnan R, Ramakers M, Claes F, Young L, Shahrooei M, Wilkinson H, Desager M, Mengistu Tadesse W, Nilsson KPR, Hammarstrom P, Aertsen A, Carpentier S, Van Eldere J, Rousseau F, Schymkowitz J (2018) Aggregating sequences that occur in many proteins constitute weak spots of bacterial proteostasis. Nat Commun 9 (1):866. doi:10.1038/s41467-018-03131-0
- Michiels E, Roose K, Gallardo R, Khodaparast L, Khodaparast L, van der Kant R, Siemons M, Houben B, Ramakers M, Wilkinson H, Guerreiro P, Louros N, Kaptein SJF, Ibanez LI, Smet A, Baatsen P, Liu S, Vorberg I, Bormans G, Neyts J, Saelens X, Rousseau F, Schymkowitz J (2020) Reverse engineering synthetic antiviral amyloids. Nat Commun 11 (1):2832. doi:10.1038/s41467-020-16721-8